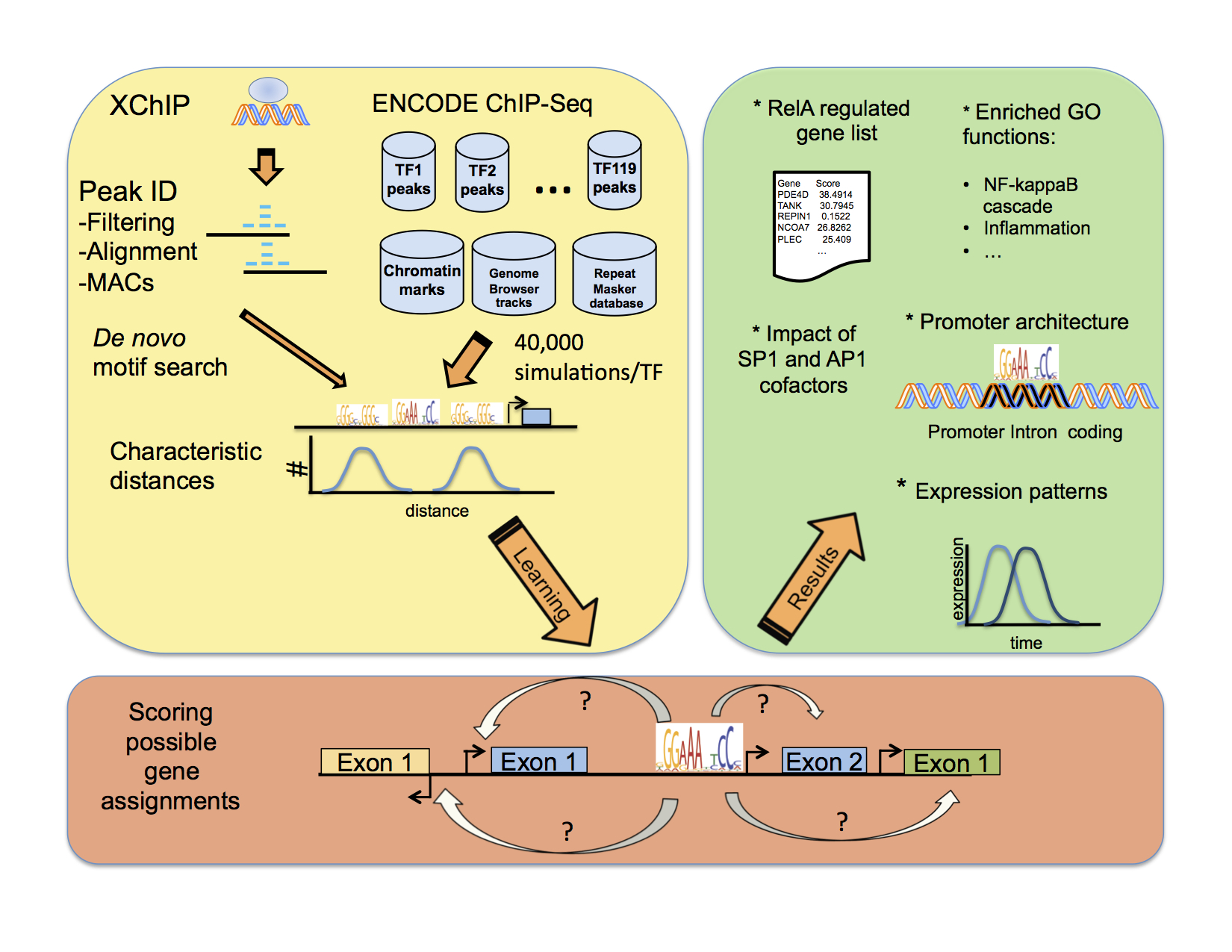
We present a detailed map of RelA promoters obtained by XChip-Seq, the motifs and co-occurences thereof. We then describe a probabilistic approach to infer the RelA regulatory network and show the newtork derived using our method. |
|
Link to Supplementary Data |
|
Visually inspect
XChiP-Seq, RelA peaks in UCSC Genome Browser. |
|
Read the paper (Oxford Journals in Nucleic Acids Research) |
|
View the data in an interactive data viewer and compute significance of correlation with your data (coming soon). |
The work has been done in Brasier Lab and the Rowicka Lab (UTMB, USA). |